Approach
The approach being pursued by the HMS LINCS Center is to expose diverse cells types to perturbations, individually or in combination, and then to assay cell responses using multiple biochemical and cell biological assays. Typically, cells grown under a variety of conditions (e.g. in full serum, in synthetic media, in the presence of ECM) are exposed to biological ligands (e.g. growth factors, cytokines), to small molecule drugs, or to combinations of perturbagens. Data on cell state (e.g. mitosis, apoptosis) or on the levels, modification state, and/or localization of signaling proteins is then collected by fixed- or live-cell microscopy or by analysis of lysates using immunoassays, reverse phase protein arrays, and/or mass spectrometry. Transcriptional responses are measured using RNA-Seq or the 1000-plex L1000 platform developed by the Broad LINCS Center. We also collect data on the perturbing agents themselves, in particular on the spectrum of kinases that are bound to or inhibited by a particular drug (recognizing that all drugs exhibit some polypharmacology). Collectively, these data are mined to derive insight into the underlying response pathways (Figure 1).
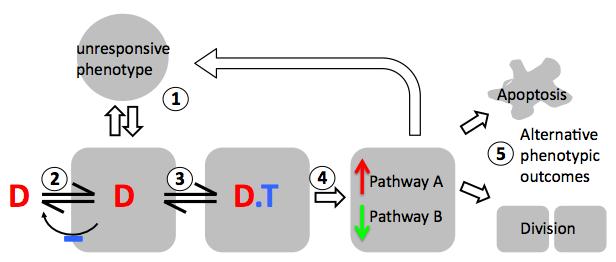
Conceptual view of a drug (D) binding to target (T) and triggering cell responses, showing elements that may vary between cell types or between drugs with different structures but similar target specificities. 1. Phenotypic variability of cells; 2. Cellular pharmacokinetics (small molecule influx, efflux, sequestration, and metabolism); 3. Drug targeting and polypharmacology; 4. Perturbation of networks in which the target protein(s) are embedded; 5. Integrated physiological responses that determine phenotypic outcome.
Adaptive Dataset Design
Given the heterogeneity and high dimensionality of the data we are collecting and the heterogeneity between cell types in responsiveness to perturbagens over a given dose and time scale, it is neither practical nor productive to achieve comprehensive coverage of the entire space of perturbations and measurements. Instead, we implement an adaptive approach in which relatively simple and inexpensive assays are performed on large numbers of cell lines/cell types and relatively complex assays are performed on subsets of cells for which preliminary data suggest the results will be most informative. This creates data subsets that represent deep looks into parts of a much larger biological space. Figure 2 provides a concrete example of this for a selected subset of HMS LINCS datasets.
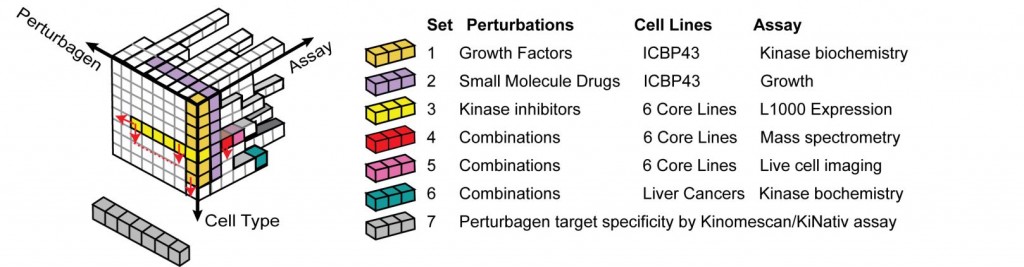
Relationship among a few specific HMS LINCS datasets (colored cubes) within the context of the complete space of possible perturbagens and assays.
Dataset Integration
Creating an effective scheme for adaptive design and integrating the resulting ‘ragged’ data structure that is progressively expanding based on emerging requirements or models is one of the primary challenges of our approach.