Experimental Methods
In addition to performing established biochemical and cell biological assays, the HMS LINCS Center participates in the development and improvement of cutting-edge experimental methods. The goal of the Center is to integrate these new methods into our adaptive data collection approach. The following list of methods developed by HMS LINCS will be updated regularly as our work progresses.
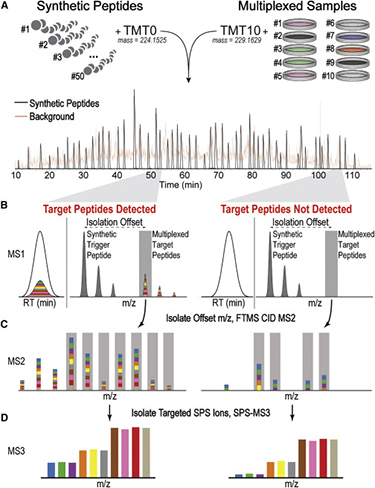
TOMAHAQ mass spectrometry
Molecular Cell (2017) 65(2):361-370. doi:10.1016/j.molcel.2016.12.005 PMID:28065596 PMCID:PMC5250569
In proteomics, two fundamental types of multiplexing are available to increase throughput. Many peptides can be targeted for quantification during a single run (peptide multiplexing), and higher-order multiplexing is possible at the sample level using a method such as isobaric labeling. Targeted mass spectrometry-based analyses have become vital for multiplexing the measurement of many peptides, but isobaric tagging-based multiplexing in targeted assays has not been demonstrated with unfractionated mixtures. Leveraging improved mass spectrometry instrumentation capable of sensitive MS3 analysis of reporter ions, we developed a 10-plex sample multiplexing strategy that enables accurate, targeted quantification directly from proteolyzed cell lysates.
Normalized growth rate inhibition (GR) metrics
Nature Methods (2016) 13(6):521-527. doi:10.1038/nmeth.3853 PMID:27135972 PMCID:PMC4887336
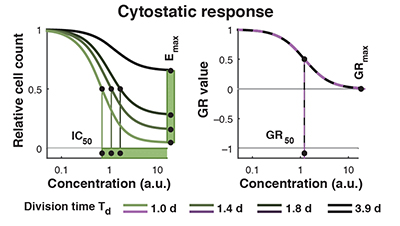 Traditional perturbagen-response metrics such as IC50, Emax, and AUC can vary dramatically, independent of the underlying biology, due to natural differences in cellular proliferation rate, variation in growth conditions, or changes in the duration of a dose-response experiment. We developed methods to parameterize growth rate data instead using normalized growth rate inhibition (GR) metrics, which yield GR50, GRmax, GRAOC, and hGR (Hill slope) values that are largely independent of cell division rate and assay duration. Importantly, only modest changes in experimental procedures are necessary to enable calculation of GR metrics from perturbagen-response datasets.
Traditional perturbagen-response metrics such as IC50, Emax, and AUC can vary dramatically, independent of the underlying biology, due to natural differences in cellular proliferation rate, variation in growth conditions, or changes in the duration of a dose-response experiment. We developed methods to parameterize growth rate data instead using normalized growth rate inhibition (GR) metrics, which yield GR50, GRmax, GRAOC, and hGR (Hill slope) values that are largely independent of cell division rate and assay duration. Importantly, only modest changes in experimental procedures are necessary to enable calculation of GR metrics from perturbagen-response datasets.
CycIF, a high-throughput cyclic immunofluorescence method
Nature Communications (2015) 6:8390. doi:10.1038/ncomms9390 PMID:26399630 PMCID:PMC4587398
Current Protocols in Chemical Biology (2016) 8(4):251-264. doi:10.1002/cpch.14 PMID:27925668 PMCID:PMC5233430
 There is an emerging need in numerous fields of biological research to increase the multiplicity of single-cell measurements. Multiplexed single-cell techniques have the potential to reveal important interdependencies between a cell’s local environment and its differentiation status, signal transduction pathway activity, and morphological phenotypes that are not evident when each feature is measured independently. To this end, we developed a robust, multiplexed immunofluorescence imaging method, named Cyclic Immunofluorescence (CycIF), using public domain chemistry and existing instruments that enables low-cost, high-dimensionality imaging assays at the single-cell level.
There is an emerging need in numerous fields of biological research to increase the multiplicity of single-cell measurements. Multiplexed single-cell techniques have the potential to reveal important interdependencies between a cell’s local environment and its differentiation status, signal transduction pathway activity, and morphological phenotypes that are not evident when each feature is measured independently. To this end, we developed a robust, multiplexed immunofluorescence imaging method, named Cyclic Immunofluorescence (CycIF), using public domain chemistry and existing instruments that enables low-cost, high-dimensionality imaging assays at the single-cell level.
A one-step, live-cell imaging assay for mitotic arrest and apoptotic state
J Biomol Screen (2013) 18 (9):1062–1071. doi:10.1177/1087057113493804 PMID:23788527 PMCID:PMC3783590
Current Protocols in Chemical Biology (2014) 6(1):1-5. doi:10.1002/9780470559277.ch130140 PMID:24652619 PMCID:PMC4016950
 Discriminating different mechanisms that compromise drug sensitivity in cells in culture requires multiplexed readout of response, which is often accomplished using mRNA profiling, multiplexed gene expression reporters, and high-content imaging assays. These assays can be highly informative but typically are costly and complex. Furthermore, it can be difficult to infer alternative mechanistic effects on drug response pathways from gene expression and other multiplex readouts where the relationship between readout and drug response pathway is complex. Here, we describe a new one-step, no-wash imaging assay that uses three dyes (Hoechst33342, LysoTracker-Red, and DEVD-NucView488) to stain living cells and enables measurement of multiple physiological changes in cells related to mitotic and apoptotic status following treatment with anti-mitotic small molecule drugs.
Discriminating different mechanisms that compromise drug sensitivity in cells in culture requires multiplexed readout of response, which is often accomplished using mRNA profiling, multiplexed gene expression reporters, and high-content imaging assays. These assays can be highly informative but typically are costly and complex. Furthermore, it can be difficult to infer alternative mechanistic effects on drug response pathways from gene expression and other multiplex readouts where the relationship between readout and drug response pathway is complex. Here, we describe a new one-step, no-wash imaging assay that uses three dyes (Hoechst33342, LysoTracker-Red, and DEVD-NucView488) to stain living cells and enables measurement of multiple physiological changes in cells related to mitotic and apoptotic status following treatment with anti-mitotic small molecule drugs.

ActivX ATP probe-enhanced, high-throughput mass spectrometry
Analytical Chemistry (2013) 85(9):4666–4674. doi:10.1021/ac303478g PMID:23607489 PMCID:PMC3771683
Kinome-wide profiling aims to provide a systematic, unbiased look at kinase levels and modification states across biological and pathological processes. Current kinase analysis methods, however, are low in throughput and often poor at detecting low-abundance kinases unless time-consuming cellular fractionation steps are undertaken. We describe here a robust method that combines ActivX ATP probe (AAP) affinity reagents (ATP analogues) with 6-plex tandem mass tag (TMT) isotopic labeling of cell lysates and enables multiplexed analysis of ~90 kinases across six conditions in a single LC-MS run.