Project Explorer
Kinase inhibitor pathways
Inhibitors of ATM: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| CP466722 | T | ✔ | |
| KU-55933 | T | ||
| KU-60019 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of AKT (AKT1, AKT2, AKT3): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A443654 | T | ✔ | ||||||||||
| AT-7867 | T | |||||||||||
| GSK690693 | I | |||||||||||
| KIN001-102 | T | ✔ |
|
|||||||||
| MK2206 | I | |||||||||||
| Triciribine | I |
|
||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of PARP (PARP, PARP-2): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| Olaparib | I | ||
| Veliparib | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of MDM2: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nutlin 3a | T |
|
||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of p90RSK: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| Fmk | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of Raf (B-Raf, C-Raf): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZ-628 | T | ✔ | ||||||||||
| GDC-0879 | T | |||||||||||
| GW-5074 | T | ✔ | ||||||||||
| HG-6-64-01 | T | ✔ | ||||||||||
| L-779450 | T |
|
||||||||||
| PLX-4720 | T | ✔ | ||||||||||
| R7204 | A | ✔ | ||||||||||
| RAF 265 | I | |||||||||||
| SB 590885 | T | ✔ | ||||||||||
| Sorafenib | A | ✔ |
|
|||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of FAK: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| PF-431396 | T | ||
| PF-562271 | T | ||
| PF-573228 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of DNA-PK: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| NU7441 | T | ✔ | |
| PI103 | T | ||
| QL-X-138 | T | ✔ | |
| SAR245409 | I | ||
| Torin2 | T | ✔ | |
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of PKC: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| Chelerythrine | T | ||
| Enzastaurin | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of CSF1R: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| HG-6-64-01 | T | ✔ | |
| KIN001-269 | T | ||
| Ki20227 | T | ||
| WZ-4-145 | T | ✔ | |
| WZ-7043 | T | ✔ | |
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of p38 (p38-alpha, p38-beta, p38-delta, p38-gamma): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| BIRB 796 | T | ||
| HG-6-64-01 | T | ✔ | |
| SB 203580 | T | ||
| SB 239063 | T | ||
| TAK-715 | T | ✔ | |
| VX-745 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of CHK1/2 (CHK1, CHK2): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZD-7762 | I | ✔ | ||||||||||
| PF-477736 | T | |||||||||||
| TCS 2312 | T |
|
||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of MEK5: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| BIX 02189 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of Wee1: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| MK1775 | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of ERBB2: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Afatinib | I |
|
||||||||||
| CP724714 | T | ✔ | ||||||||||
| Lapatinib | A |
|
||||||||||
| Neratinib | I | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of Src: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bosutinib | A |
|
||||||||||
| Dasatinib | A | |||||||||||
| Saracatinib | I | |||||||||||
| WH-4-025 | T | ✔ | ||||||||||
| XMD16-144 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of IGF1R: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AEW541 | T | |||||||||||
| AG1024 | T |
|
||||||||||
| GSK1838705 | T |
|
||||||||||
| GSK1904529A | T | |||||||||||
| OSI-906 | I | |||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of HSP90 (HSP90 alpha, HSP90 beta): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17-AAG | T |
|
||||||||||
| Geldanamycin | T |
|
||||||||||
| NVP-AUY922 | T | |||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of PI3K (PI3K-alpha, PI3K-beta, PI3K-delta, PI3K-gamma): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A66 | T | |||||||||||
| AS-252424 | T |
|
||||||||||
| AS-605240 | T | |||||||||||
| AZD-6482 | T | ✔ | ||||||||||
| BEZ235 | I |
|
||||||||||
| BYL719 | T | |||||||||||
| CAL-101 | I | |||||||||||
| GDC-0941 | T | ✔ | ||||||||||
| GDC-0980 | T | |||||||||||
| GSK1059615 | I | ✔ |
|
|||||||||
| GSK1487371 | T | |||||||||||
| GSK2119563 | T |
|
||||||||||
| GSK2126458 | I | ✔ |
|
|||||||||
| NVP-BKM120 | I | |||||||||||
| SAR245408 | I | |||||||||||
| SAR245409 | I | |||||||||||
| TGX221 | T |
|
||||||||||
| Torin2 | T | ✔ | ||||||||||
| ZSTK474 | I | |||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of mTOR: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZD-8055 | I | |||||||||||
| GDC-0980 | T | |||||||||||
| GSK1059615 | I | ✔ |
|
|||||||||
| GSK2126458 | I | ✔ |
|
|||||||||
| OSI-027 | T | |||||||||||
| QL-X-138 | T | ✔ | ||||||||||
| RAD001 | T | |||||||||||
| SAR245409 | I | |||||||||||
| Sirolimus | A |
|
||||||||||
| Temsirolimus | A |
|
||||||||||
| Torin1 | T | ✔ | ||||||||||
| Torin2 | T | ✔ | ||||||||||
| WYE-125132 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of JAK (JAK1, JAK2, JAK3): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| AZD-1480 | I | ||
| CYT387 | I | ✔ | |
| KIN001-055 | T | ||
| QL-X-138 | T | ✔ | |
| Ruxolitinib | A | ||
| TG 101348 | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of MEK1/2 (MEK1, MEK2): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARRY-424704 | I | |||||||||||
| BMS-777607 | I | |||||||||||
| CI-1040 | T | |||||||||||
| GSK1120212 | I |
|
||||||||||
| PD-325901 | I | ✔ | ||||||||||
| PD-98059 | T |
|
||||||||||
| Selumetinib | I |
|
||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of c-Met: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| Amuvatinib | I | ||
| Cabozantinib | I | ||
| Crizotinib | A | ✔ | |
| Foretinib | I | ||
| JNJ38877605 | I | ||
| KIN001-237 | T | ||
| MGCD265 | I | ||
| PF-04217903 | I | ||
| PHA-665752 | T | ||
| PKI-SU11274 | T | ||
| SGX523 | I | ||
| Tivantinib | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of ALK (ALK2, ALK3, ALK5): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CH5424802 | I | |||||||||||
| D 4476 | T | |||||||||||
| GSK1838705 | T |
|
||||||||||
| LDN-193189 | T | |||||||||||
| NVP-TAE684 | T | |||||||||||
| SB 525334 | T | |||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of JNK (JNK1, JNK2, JNK3): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AS-601245 | T | ✔ | ||||||||||
| CC-401 | I | |||||||||||
| CG-930 | T | |||||||||||
| JNK-9L | T | ✔ | ||||||||||
| JNK-IN-5A | T | ✔ |
|
|||||||||
| SP600125 | T | |||||||||||
| ZG-10 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of p53: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| JNJ26854165 | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of PLK (PLK1, PLK2, PLK3): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BI-2536 | I | ✔ | ||||||||||
| GSK461364 | I | ✔ |
|
|||||||||
| GW843682 | T | ✔ | ||||||||||
| HMN-214 | T | |||||||||||
| NPK76-II-72-1 | T | ✔ | ||||||||||
| ON-01910 | I | |||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of c-Abl (c-Abl, Bcr-Abl): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| GNF2 | T | ||
| HG-5-113-01 | T | ✔ | |
| HG-6-64-01 | T | ✔ | |
| Imatinib | A | ||
| Nilotinib | A | ||
| Ponatinib | I | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of Aurora (AURKA, AURKB, AURKC): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Barasertib | I | ✔ | ||||||||||
| GSK1070916 | T |
|
||||||||||
| JWE-035 | T | ✔ | ||||||||||
| KIN001-220 | T | ✔ | ||||||||||
| MLN8054 | T | ✔ | ||||||||||
| Tozasertib | I |
|
||||||||||
| XMD16-144 | T | ✔ | ||||||||||
| ZM-447439 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of EGFR: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Afatinib | I |
|
||||||||||
| Canertinib | I | |||||||||||
| Erlotinib | A |
|
||||||||||
| Gefitinib | A |
|
||||||||||
| HG-5-88-01 | T | ✔ | ||||||||||
| HG-6-64-01 | T | ✔ | ||||||||||
| Lapatinib | A |
|
||||||||||
| Pelitinib | I | |||||||||||
| Tyrphostin | T | ✔ |
|
|||||||||
| WZ-4-145 | T | ✔ | ||||||||||
| WZ-4002 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of AMPK (AMPK-alpha1, AMPK-alpha2): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| A769662 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of CDK (CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, CDK9): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AT-7519 | I | |||||||||||
| AZD-5438 | I | |||||||||||
| BMS-387032 | I | |||||||||||
| Fascaplysin | T |
|
||||||||||
| Flavopiridol | I | |||||||||||
| KIN001-270 | T | |||||||||||
| MLS000911536 | T | ✔ | ||||||||||
| NU6102 | T |
|
||||||||||
| PD0332991 | I | ✔ | ||||||||||
| PHA-793887 | T | |||||||||||
| Purvalanol A | T |
|
||||||||||
| Seliciclib | I | |||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of PDK1: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| BX-912 | T | ✔ | |
| KIN001-244 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of Bcl-2: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| ABT-737 | I | ||
| Docetaxel | A | ||
| Paclitaxel | A | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of FLT3: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC220 | I | |||||||||||
| Dovitinib | I | |||||||||||
| HG-6-64-01 | T | ✔ | ||||||||||
| KW2449 | I | |||||||||||
| Lestaurtinib | I |
|
||||||||||
| Linifanib | I | |||||||||||
| Sunitinib | A |
|
||||||||||
| WZ-3105 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of MNK (MNK1, MNK2): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| QL-X-138 | T | ✔ | |
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of ERK5: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| LRRK2-in-1 | T | ✔ | |
| XMD8-85 | T | ✔ | |
| XMD8-92 | T | ✔ | |
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of GSK3 (GSK3A, GSK3B): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| CHIR-99021 | T | ||
| KIN001-042 | T | ||
| KIN001-043 | T | ||
| SB 216763 | T | ||
| TWS119 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of FGFR (FGFR1, FGFR3): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BGJ398 | I | |||||||||||
| HG-6-64-01 | T | ✔ | ||||||||||
| PD-173074 | T | ✔ |
|
|||||||||
| Vargatef | I | |||||||||||
| WZ-7043 | T | ✔ | ||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of EPHB3: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| XMD14-99 | T | ✔ | |
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of IKK (IKK-alpha, IKK-beta, IKK-epsilon): |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMS-345541 | T | ✔ | ||||||||||
| TPCA-1 | T |
|
||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of c-Kit: |
||||||||||||
| Name | C* | KS* | Signature [GI50, log10(M)] ** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amuvatinib | I | |||||||||||
| HG-6-64-01 | T | ✔ | ||||||||||
| Masitinib | I | |||||||||||
| OSI-930 | I | |||||||||||
| Sunitinib | A |
|
||||||||||
|
|
||||||||||||
|
||||||||||||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
** Data from Heiser et. al 2012
Inhibitors of DDR1: |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| ALW-II-38-3 | T | ✔ | |
| ALW-II-49-7 | T | ✔ | |
| QL-XI-92 | T | ✔ | |
| WZ-4-145 | T | ✔ | |
| WZ-7043 | T | ✔ | |
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
Inhibitors of ERK1/2 (ERK1, ERK2): |
|||
| Name | C* | KS* | Signature [GI50, log10(M)] |
|---|---|---|---|
| FR180204 | T | ||
* C = classification (Approved/Investigational/Tool);
KS =
KinomeScan profile available (links to dataset)
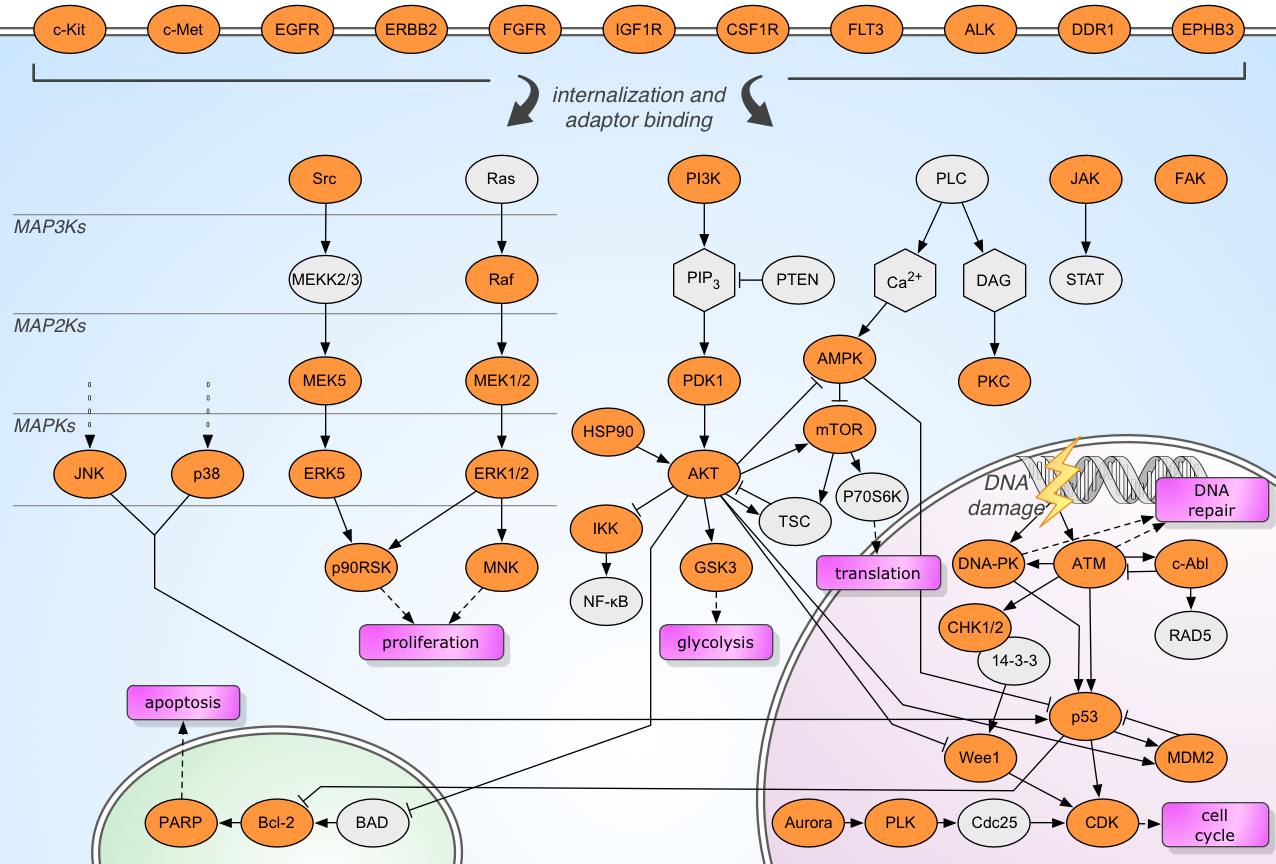
One component of the HMS LINCS initiative is the creation of a highly curated library of kinase inhibitors. We are focusing on clinical compounds (FDA approved or undergoing trials) and tool compounds which are highly selective for important kinases.
This network map of key signal transduction pathways includes the majority of kinases we can selectively target with small molecules, as well as information on the relevant inhibitors in the HMS LINCS library. This allows biologists to find compounds which target a given kinase or affect a specific phenotypic outcome, as well as to identify compounds with potentially similar or synergistic effects by virtue of targeting different nodes in the same signal transduction cascade.
Instructions
Hover or tap on a kinase shaded in orange to view a box listing compounds in the HMS LINCS library which inhibit the activity of that kinase. For each compound, the box shows the following summary information: the inhibitor's classification (Approved/Investigational/Tool - see below), a link to a KinomeScan profile of the compound (if available), and a graphical visualization of the compound's ability to inhibit growth of a panel of six breast cancer cell lines (data from Heiser et. al 2012). Protein, compound and cell line names are all hyperlinked to entries in the HMS LINCS Database which contain detailed information.
Compound classifications:
- Approved: FDA approved for use in human subjects
- Investigational: Undergoing clinical trials
- Tool: Only suitable for research in cell culture
You may also use our full database search engine to find compounds or protein targets by name or other attributes.
References
- Heiser, L. et al. (2012) Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci. 109(8), 2724-9. doi:10.1073/pnas.1018854108 PMID:22003129 PMCID:PMC3286973