Publication Summary
A Multi-center Study on the Reproducibility of Drug-Response Assays in Mammalian Cell Lines
Mario Niepel*,1,¶, Marc Hafner*,1,†, Caitlin E. Mills*,1, Kartik Subramanian1, Elizabeth H. Williams1, Mirra Chung1, Benjamin Gaudio1, Anne Marie Barrette2, Alan D. Stern2, Bin Hu2, James E. Korkola3, LINCS Consortium, Joe W. Gray3, Marc R. Birtwistle2,§,††, Laura M. Heiser3,††,α, and Peter K Sorger1,††
*These authors contributed equally
††Co-corresponding authors
αLead contact
1HMS LINCS Center, Laboratory of Systems Pharmacology, Department of Systems Biology, Harvard Medical School, Boston, MA 02115, USA
2Drug Toxicity Signature Generation (DToxS) LINCS Center, Department of Pharmacological Sciences, Mount Sinai Institute for Systems Biomedicine, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place Box 1215, New York NY 10029
3Microenvironment Perturbagen (MEP) LINCS Center, Oregon Health & Sciences University, Robertson Life Sciences Building (RLSB) – CL3G, 2730 SW Moody Avenue, Portland, OR 97201
¶Current address: Ribon Therapeutics, Inc. 99 Hayden Avenue, Building D, Suite 100, Lexington, MA 02421
†Current address: Department of Bioinformatics & Computational Biology, Genentech, Inc., South San Francisco, CA 94080
§Current address: Clemson University, Dept. of Chemical and Biomolecular Engineering, Earle Hall, 206 S. Palmetto Blvd., Clemson, SC 29634
Cell Syst. 9(1):35-48.e5.
doi:10.1016/j.cels.2019.06.005 PMID:31302153
Synopsis
A major focus of the LINCS program has been the design and implementation of FAIR (Findable, Accessible, Interoperable, and Reusable) data practices. This paper investigates the reproducibility of experiments measuring the sensitivity of MCF 10A epithelial mammary cells to small molecule drugs. Initial “replicate” experiments at five LINCS centers supplied with identical cell, media, and drug aliquots yielded up to 200-fold variation in sensitivity measurements; analysis of sources of irreproducibility led to improvements in experimental and analytical procedures, such that that technical staff without previous exposure to our protocol and trained two years after the start of the study could obtain results indistinguishable from assays performed two years previously.
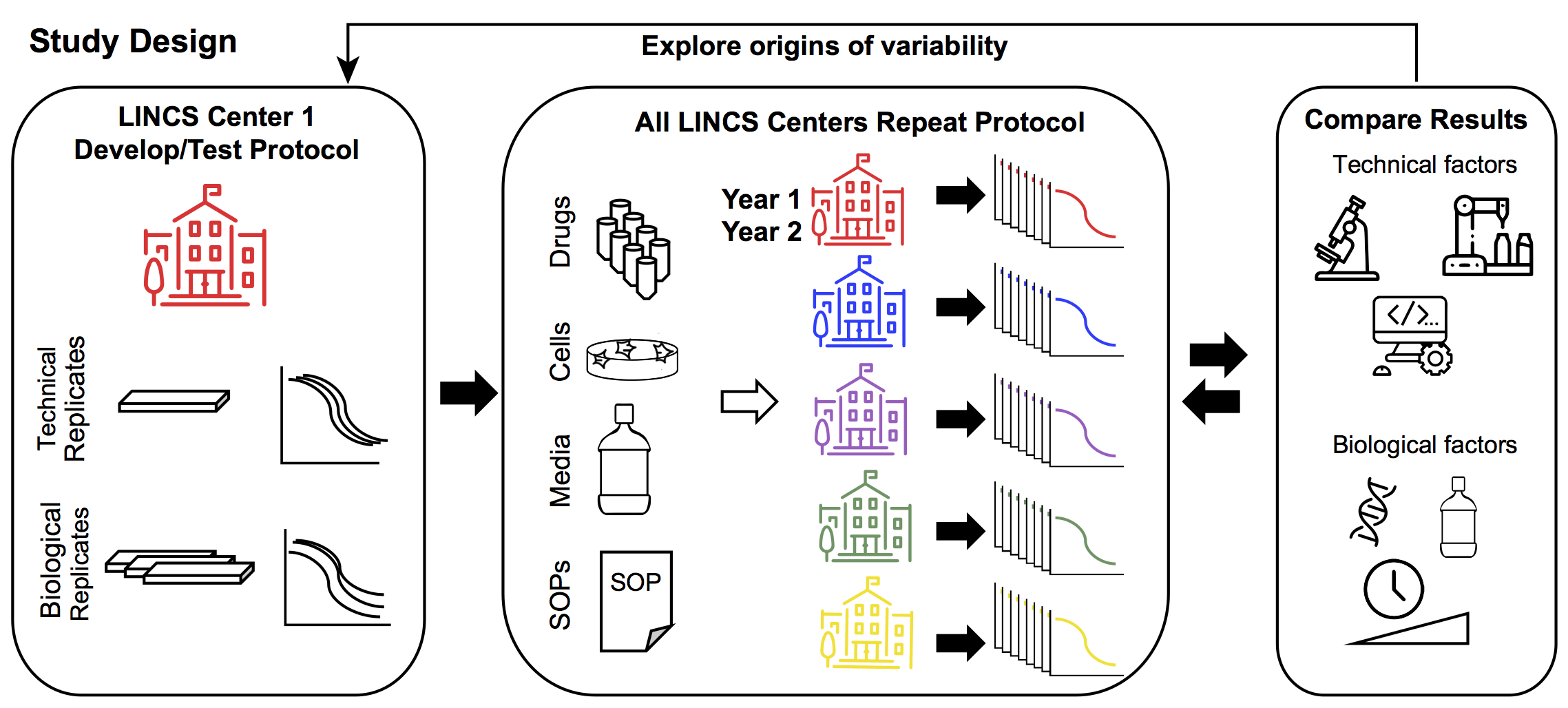
LINCS Center One defined the experimental protocol and established within-Center reproducibility by assessment of technical (different wells, plates, same day) and biological (different days) replicates. Common stocks of drugs, cells, and media, as well as a standard experimental protocol was distributed to each of the five data generation centers. Each center performed 72h dose-response measurements for each of the 8 drugs. LINCS Center One explored the various technical and biological drivers of variability. Technical drivers of variability include assay read-out, use of automation, and analytical pipelines. Biological drivers of variability include cell line isolate, reagents, assay duration, and dose range. This information was fed back to the other Centers to refine their dose-response measurements.
Key Findings
- Our results show that irreproducibility arises from unexpected interplay between experimental protocol and true biological variability and contradict the belief that counting cells is such a simple procedure that different assays can be substituted for each other without consequence (i.e. CellTiter-Glo for direct counts).
- Sources of technical variation included inappropriate assay substitution (direct cell counts versus CellTitre-Glo), edge effects and non-uniform cell growth across microtitre plates (reduced through randomized plate layouts), human pipetting error (reduced by automation), dose range and duration of exposure, and differences in data handling (potentially mitigated by dockerized data analysis pipelines).
- In contrast to studies emphasizing the role of genetic instability in irreproducibility, the effect of isolate-to-isolate differences on drug response assays was negligible compared to the ways in which drugs and cells were plated into multi-well plates and counted.
Abstract
Evidence that some influential biomedical results cannot be repeated has increased interest in practices that generate data meeting findable, accessible, interoperable and reproducible (FAIR) standards. Multiple papers have identified examples of irreproducibility, but practical steps for increasing reproducibility have not been widely studied. Here, seven research centers in the NIH LINCS Program Consortium investigate the reproducibility of a prototypical perturbational assay: quantifying the responsiveness of cultured cells to anti-cancer drugs. Such assays are important for drug development, studying cell biology, and patient stratification. While many experimental and computational factors have an impact on intra- and inter-center reproducibility, the factors most difficult to identify and correct are those with a strong dependency on biological context. These factors often vary in magnitude with the drug being analyzed and with growth conditions. We provide ways of identifying such context-sensitive factors, thereby advancing the conceptual and practical basis for greater experimental reproducibility.
Available data and software
Data analyses available via an on-line set of Jupyter notebooks (https://github.com/labsyspharm/MCF10A_DR_reproducibility and https://github.com/datarail/datrail”). A list of best practices as well as an introduction to GR metrics can be found at http://www.grcalculator.org. Center 1 datasets are listed below. Data from centers 2-5 as well as mean values for all centers are available through https://www.synapse.org/#!Synapse:syn18456348/.
| Figure | Associated Dataset | Synapse ID | HMS LINCS Dataset |
|---|---|---|---|
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist A, 2017. Normalized growth rate inhibition values. | syn18456349 | Pending | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist A, 2019. Normalized growth rate inhibition values. | syn18456350 | 20278-20283 | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist B. Normalized growth rate inhibition values. | syn18456351 | Pending | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist C. Normalized growth rate inhibition values. | syn18456352 | Pending | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist A, 2017. GR metrics. | syn18483759 | 20284-20286 | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist A, 2019. GR metrics. | syn18483760 | Pending | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist B. GR metrics. | syn18483761 | Pending | |
| Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Repeat performed at Center 1 by Scientist C. GR metrics. | syn18483762 | ||
| Rolling-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Normalized growth rate inhibition values for biological replicate 1. | syn18478972 | 20318 | |
| Rolling-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Normalized growth rate inhibition values for biological replicate 2. | syn18478973 | 20319 | |
| Rolling-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Normalized growth rate inhibition values for biological replicate 3. | syn18478974 | 20320 | |
| Rolling-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. GR metrics for biological replicate 1. | syn18478975 | 20321 | |
| Rolling-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. GR metrics for biological replicate 2. | syn18478976 | 20322 | |
| Rolling-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. Fixed-time-point sensitivity measures of the MCF 10A breast cell line to 8 small molecule perturbagens. GR metrics for biological replicate 3. | syn18478977 | 20323 |
Funding Sources
This work was funded by grants U54-HL127365 to PKS; U54-HG008098 to RI, MRB, and EAS; R01- GM104184 to MRB; U54HL127624 to MM and AM; U54-HG008100 to JWG, LMH, and JEK; U54- HG008097 to JDJ; U54-NS091046 to CNS; U54-HL127366 to TRG and AD. ADS and AMB were supported by a NIGMS-funded Integrated Pharmacological Sciences Training Program grant T32GM062754.