Publication Summary
Multi-omics profiling establishes the polypharmacology of FDA Approved CDK4/6 inhibitors and the potential for differential clinical activity
Marc Hafner*,1,3, Caitlin Elizabeth Mills*,1, Kartik Subramanian1, Chen Chen1, Mirra Chung1, Sarah A. Boswell1, Robert A. Everley1, Changchang Liu1, Charlotte Sanden Walmsley2, Dejan Juric1,2,ɸ, and Peter Karl Sorger1,†,ɸ
*These authors contributed equally to this work.
ɸThese authors contributed equally to this work.
1Laboratory of Systems Pharmacology, Department of Systems Biology, Harvard Medical School, Boston, MA 02115
2Termeer Center for Targeted Therapies, Massachusetts General Hospital Cancer Center, Boston, MA 02114
3Current address: Department of Bioinformatics & Computational Biology, Genentech, Inc., South San Francisco, CA 94080
† Lead contact: Peter Sorger (peter_sorger [at] hms.harvard.edu); orcid.org/0000-0002-3364-1838 copying sorger_admin [at] hms.harvard.edu).
Cell Chem Biol. 26(8):1067-1080.e8.
doi:10.1016/j.chembiol.2019.05.005 PMID:31178407 PMCID:PMC6936329
Synopsis
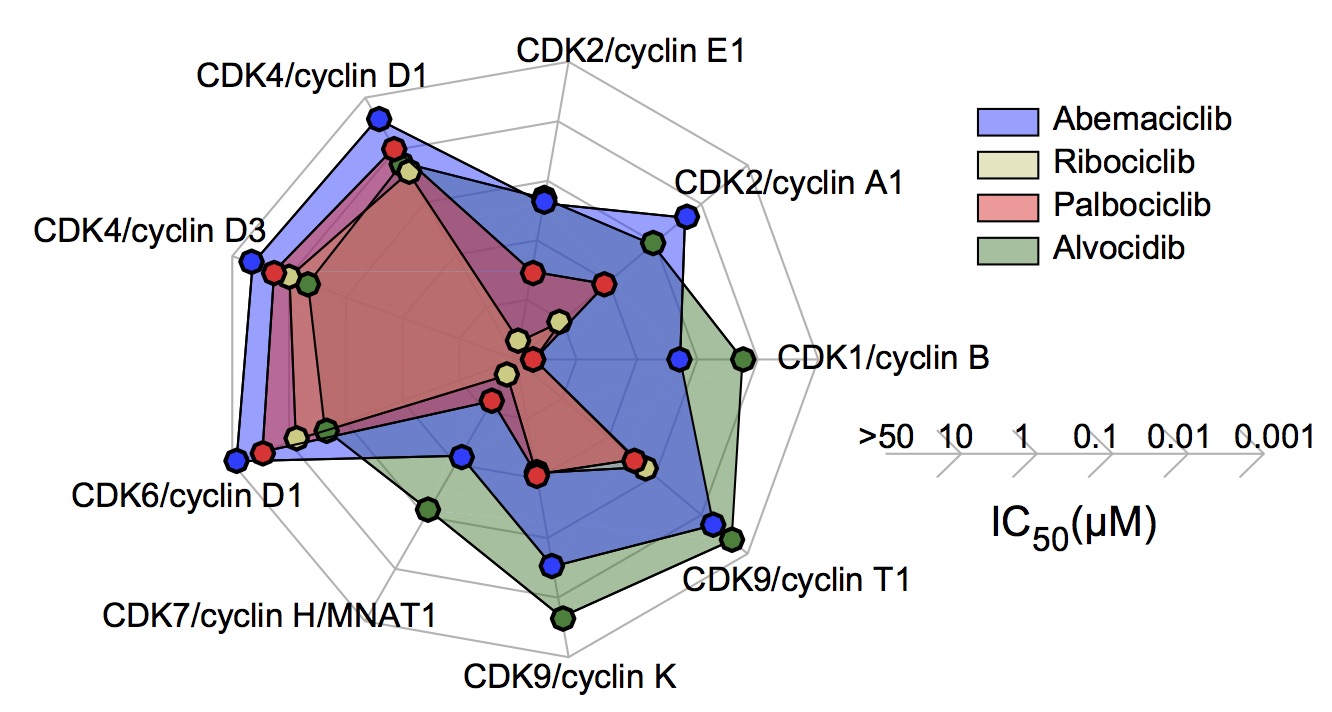
Figure 3e. IC50 values for CDK/cyclin complexes for CDK4/6 inhibitors and alvocidib as
measured using purified kinases in vitro.
Over-expression of cyclin-dependent kinase (CDK) proteins, leading to dysregulation of the cell cycle, is common in solid tumors including breast cancer. Drugs that inhibit CDK4 and CDK6, including palbociclib (Ibrance®), ribociclib (KISQALI®) and abemaciclib (Verzenio®), are currently regarded as some of the most promising new treatments for hormone receptor-positive breast cancer. While emerging evidence suggests that these drugs differ in the clinic, their target profiles and biological activities have not been directly compared. We analyzed palbociclib, ribociclib, and abemaciclib using five complementary experimental approaches and found that they have substantial differences in biological activities and secondary targets in breast cancer cell lines. Multiple lines of evidence suggest that inhibition of additional CDK proteins by abemaciclib may be therapeutically advantageous.
Key Findings
Abstract
The target profiles of many drugs are established early in their development and are not systematically revisited at the time of FDA approval. Thus, it is often unclear whether therapeutics with the same nominal targets but different chemical structures are functionally equivalent. In this paper we use five different phenotypic and biochemical assays to compare approved inhibitors of cyclin-dependent kinases 4/6 – collectively regarded as breakthroughs in the treatment of hormone receptor-positive breast cancer. We find that transcriptional, proteomic and phenotypic changes induced by palbociclib, ribociclib, and abemaciclib differ significantly; abemaciclib in particular has advantageous activities partially overlapping those of alvocidib, an older polyselective CDK inhibitor. In cells and mice, abemaciclib inhibits kinases other than CDK4/6 including CDK2/Cyclin A/E ¬– implicated in resistance to CDK4/6 inhibition – and CDK1/Cyclin B. The multi-faceted experimental and computational approaches described here therefore uncover under-appreciated differences in CDK4/6 inhibitor activities with potential importance in treating human patients.
Datasets and Associated Metadata and Code
The following data are available for download.
| Table | Description | HMS LINCS ID | Synapse ID | Other Resources |
|---|---|---|---|---|
| S1 | DGE-seq Gene expression data for 7 cell lines treated with abemaciclib, palbociclib, ribociclib at 0.1, 0.3, 1, or 3 μM, or alvocidib at 0.1 or 1 μM for 6 hours. | syn18488088 | GEO GSE125215 | |
| S1 | Enrichment analysis of genes in signature 1 (CDK4/6 inhibitor signature) and signature 2 (abemaciclib-specific signature). | syn18488087 | ||
| S1 | mRNA-seq gene expression data for seven cell lines treated with one of three CDK4/6 inhibitors at 0.3, 1.0, or 3.0 μM for 6 or 24 hours. | syn18488086 | GEO GSE99116 | |
| S1 | mRNA-seq gene expression data for MCF7 xenografts treated with ribociclib, palbociclib, abemaciclib or vehicle. | syn18488096 | GEO GSE124854 | |
| S2 | Kinase set library. | syn18508419 | ||
| S2 | Inferred differential kinase activity for MCF7 cells treated with palbociclib or abemaciclib at either 0.3 or 3.0 μM. | syn18488090 | ||
| S2 | Differentially expressed phosphopeptides in MCF7 cells treated with palbociclib or abemaciclib at either 0.3 or 3.0 μM and untreated controls. | syn18488089 | ||
| S3 | KINOMEscan results for ribociclib, palbociclib, or abemaciclib at 0.1 and 1.0 μM. | 20330, 20339, 20338 | syn18488091 | |
| S4 | Differentially inhibited kinases in lysates treated with ribociclib, palbociclib or abemaciclib at 0.1, 1, or 10 μM and untreated controls measured by MIB/MS. | syn18488092 | ||
| S5 | IC50 values for inhibition or binding of kinases by ribociclib, palbociclib, abemaciclib, and alvocidib. | syn18488093 | ||
| S6 | GR metrics and biphasic fitting parameters for the response of 34 breast cancer cell lines to palbociclib and abemaciclib. | 20344 | syn18488095 | |
| S6 | GR values and increased fraction of dead cells for the response of MCF7 and Hs 578T parental and 1μM-palbociclib adapted cells to ribociclib, palbociclib, and abemaciclib. | syn18488098 | ||
| S6 | GR values and increased fraction of dead cells for the response of patient-derived MGH312 cells to ribociclib, palbociclib, and abemaciclib. | syn18488099 | ||
| S6 | GR values and increased fraction of dead cells for the response of 34 breast cancer cell lines to palbociclib and abemaciclib. | 20343 | syn18488094 | |
| S6 | Time-dependent GR values and increased fraction of dead cells for the response of MCF7, Hs 578T, and PDX12-58 cells to ribociclib, palbociclib, and abemaciclib. | syn18488097 |
Funding Sources
This work was funded by P50-GM107618, U54-CA225088 and U54-HL127365 grants awarded to Peter K. Sorger and Dejan Juric.