Publication Summary
Systematic analysis of BRAFV600E melanomas reveals a role for JNK/c-Jun pathway in adaptive resistance to drug-induced apoptosis
Mohammad Fallahi-Sichani1, Nathan J. Moerke1, Mario Niepel1, Tinghu Zhang2,3, Nathanael S. Gray2,3, and Peter K. Sorger1
1 HMS LINCS Center, Harvard Medical School, Boston, MA; 2 Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA; 3 Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA
Mol Syst Biol (2015) 11: 797
doi:10.15252/msb.20145877 / PMID:25814555 / PMCID:PMC4380931
Synopsis
Adaptive responses to RAF/MEK inhibitors are analyzed systematically across a panel of BRAFV600E melanoma lines to reveal a role for cell-to-cell variability induced by the JNK/c-Jun pathway and other factors in adaptive drug resistance.
Key Findings
- Adaptive responses are profiled using a combination of multiplex measurements across time, dose, cell line and drug type, statistical modeling and single-cell analysis.
- BRAFV600E melanoma lines differ in sensitivity to RAF/MEK inhibition with respect to both IC50 and maximal effect (Emax), reflecting cell-to-cell variability in drug response.
- Adaptive responses to RAF/MEK inhibition are diverse and involve multiple signaling pathways.
- The JNK/c-Jun pathway is a common adaptive response that decreases drug maximum effect.
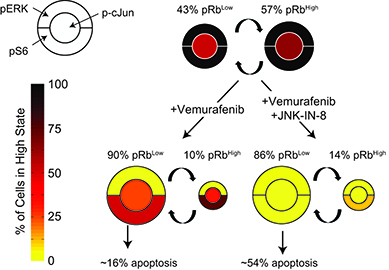
- JNK inhibition prevents induction of quiescence by RAF inhibition and promotes apoptosis.
Abstract
Drugs that inhibit RAF/MEK signaling, such as vemurafenib, elicit profound but often temporary anti-tumor responses in patients with BRAFV600E melanoma. Adaptive responses to RAF/MEK inhibition occur on a time scale of hours to days, involve homeostatic responses that reactivate MAP kinase signaling and compensatory mitogenic pathways and attenuate the anti-tumor effects of RAF/MEK inhibitors. We profile adaptive responses across a panel of melanoma cell lines using multiplex biochemical measurement, single-cell assays and statistical modeling and show that adaptation involves at least six signaling cascades that act to reduce drug potency (IC50) and maximal effect (i.e. Emax <<1). Among these cascades we identify a role for JNK/c-Jun signaling in vemurafenib adaptation and show that RAF and JNK inhibitors synergize in cell killing. This arises because JNK inhibition prevents a subset of cells in a cycling population from becoming quiescent upon vemurafenib treatment, thereby reducing drug Emax. Our findings demonstrate the breadth and diversity of adaptive responses to RAF/MEK inhibition and a means to identify which steps in a signaling cascade are most predictive of phenotypic response.
Explore the data
We encourage readers to explore the data and findings of this study through the associated project exploration website. To promote independent analysis, we also provide access through the links shown below to much of the raw data and model parameter data for this study.
Available Data and Signatures
| Data | Viability and apoptosis in BRAF(V600E/D) melanoma cell lines monitored by imaging (HMS Dataset #20217) | Details | Download (.xls) |
| Data | Phosphorylation state and protein levels measured in BRAF(V600E/D) melanoma cell lines monitored by Reverse Phase Protein Arrays (RPPA)(HMS Dataset #20218) | Details | Download (.xls) |
| Data | Phosphorylation state and protein levels measured in BRAF(V600E/D) melanoma cell lines monitored by imaging (HMS Dataset #20219) | Details | Download (.xls) |
| Signature | PLSR model loadings (pMEK and pERK included) from analysis of the covariation of molecular signals with cell viability and apoptosis fraction in BRAF(V600E/D) melanoma cell lines (HMS Dataset #20229) | Details | Download (.xls) |
| Signature | PLSR model loadings (pMEK and pERK excluded) from analysis of the covariation of molecular signals with cell viability and apoptosis fraction in BRAF(V600E/D) melanoma cell lines (HMS Dataset #20230) | Details | Download (.xls) |
| Signature | Average variable importance in the projection (VIP) scores from the PLSR models analyzing the covariation of molecular signals with cell viability and apoptosis fraction in BRAF(V600E/D) melanoma cell lines (HMS Dataset #20231) | Details | Download (.xls) |
| Software | A link to our HMS LINCS GitHub for access to the code for the data visualization tools presented on the associated project exploration website. | hmslincs at GitHub |
Funding Sources
NIH LINCS grant U54 HG006097 and its continuation U54 HL127365, NIH grants CA139980 to PKS and CA130876 to NSG, and a Merck Fellowship of the Life Sciences Research Foundation to MF-S.