Publication Summary
Drug adaptation influences cardiotoxicity caused by tyrosine kinase inhibitors in iPSC-derived human cardiomyocytes
Huan Wang1,*,†, Robert P. Sheehan1, Adam C. Palmer1, Robert A. Everley1, Sarah A. Boswell1, Noga Ron-Harel2, Alison E. Ringel2, Kristina M. Holton3, Connor A. Jacobson1, Alison R. Erickson1, Laura Maliszewski1, Marcia C. Haigis2, Peter K Sorger1,†,‡
1 Laboratory of Systems Pharmacology; Department of Systems Biology, Harvard Medical School; Boston, Massachusetts 02115, USA
2 Department of Cell Biology, Harvard Medical School; Boston, Massachusetts 02115, USA
3 Research Computing, Harvard Medical School; Boston, Massachusetts 02115, USA
* Current Address: Huan Wang, Institute of Systems Biomedicine, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 10091, China, huan_sharon_wang [at] pku.edu.cn
† Co-corresponding authors: huan_sharon_wang [at] pku.edu.cn, peter_sorger [at] hms.harvard.edu
‡ Lead contact
Cell Systems (2019) pii: S2405-4712(19)30079-1
doi:10.1016/j.cels.2019.03.009 PMID:31078528
Synopsis
Cardiotoxicity induced by anti-cancer drugs, including some already approved for clinical use, is a growing concern as the the number of cancer patients with stable disease increases, revealing the long-term cardiotoxic effects of some anti-cancer treatments. In conjunction with the Harvard Medical School Center for Cancer Systems Pharmacology (NIH U54-CA225088), HMS LINCS assessed cardiotoxicity induced by four tyrosine kinase inhibitors (Sunitinib, Sorafenib, Lapatinib and Erlotinib) in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). Our systematic profiling of hiPSC-CM gene and protein expression and physiology revealed diverse molecular responses to TKIs including adaptive drug resistance to Sorafenib. Increased understanding of TKI-induced cardiotoxicity could eventually lead to mitigation through changed dosing strategies and co-drugging with cardioprotective agents.
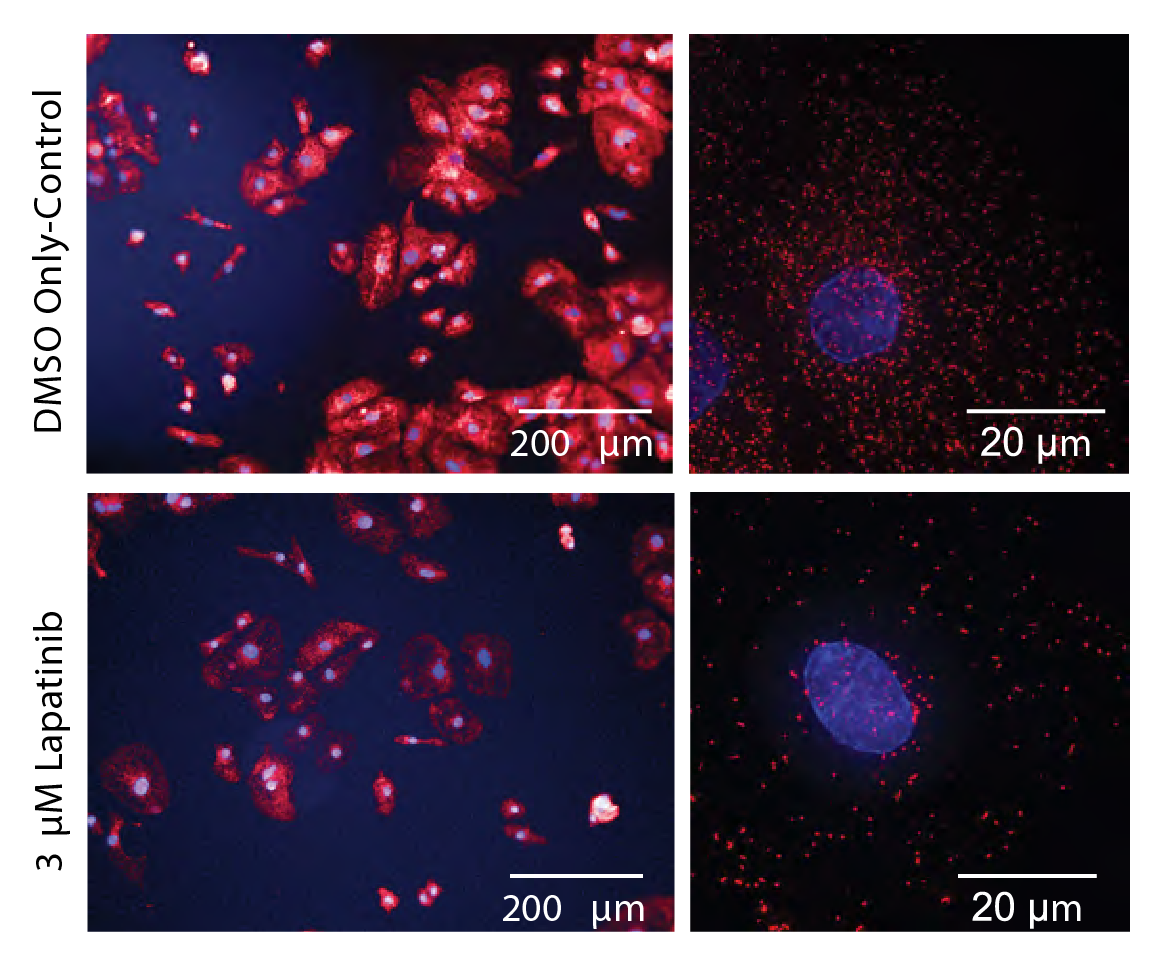
Immunofluorescent microscopy with peroxisomal staining. Proteomics revealed that 18 proteins involved in peroxisome function and biogenesis were downregulated 2-fold or more after exposure to Lapatinib, while immunofluorescent microscopy with antibodies targeting one of these proteins (PEX14) revealed an ~8-fold reduction in peroxisome number. As cardiac cells are particularly dependent on peroxisome-based oxidation of fatty acids, such reductions in peroxisome number in hiPSC-CMs is likely to impose a substantial metabolic burden.
Key Findings
- We found that cumulative drug dose and time of exposure, which are often ignored in genome-wide profiling studies, determined the magnitude of changes in gene expression.
- TKI exposure generally led to mis-regulation of processes associated with metabolism, biosynthesis and remodeling the extracellular environment.
- Latapitinib exposure reduced abundance of peroxisomes as well as proteins involved in peroxisome function and biogenesis while Sunitinib and Sorafenib exposure led to reduced spontaneous beating and expression of a key sarcomere component.
- Sorafenib exposure also reduced mitochondrial ATP production; hiPSC-CMs compensated by up-regulating glycolysis. These effects are largely reversible upon Sorafenib withdrawal.
Abstract
Tyrosine kinase inhibitors (TKIs) are widely used to treat solid tumors but can be cardiotoxic. The molecular basis for this toxicity and its relationship to therapeutic mechanisms remain unclear; we therefore undertook a systems-level analysis of human cardiomyocytes exposed to four TKIs. Cardiomyocytes (CMs) differentiated from human induced pluripotent stem cells (hiPSCs) were exposed to sunitinib, sorafenib, lapatinib or erlotinib and responses assessed by functional assays, microscopy, RNA sequencing and mass spectrometry (GEO GSE114686; PRIDE PXD012043). TKIs have diverse effects on hiPSC-CMs distinct from inhibition of tyrosine-kinase mediated signal transduction; cardiac metabolism is particularly sensitive. Following Sorafenib treatment, oxidative phosphorylation is down-regulated, resulting in a profound defect in mitochondrial energetics. Cells adapt by upregulating aerobic glycolysis. Adaptation makes cells less acutely sensitive to Sorafenib, but may have long-term negative consequences. Thus, cardiomyocytes exhibit adaptive responses to anti- cancer drugs conceptually similar to those previously shown in tumors to mediate drug resistance.
Datasets and Associated Metadata and Code
RNA-seq data were deposited in NCBI GEO under ID GSE114686.
Proteomics data were deposited in the PRIDE Archive under ID PXD012043 and will be made available upon publication of the related manuscript as per PRIDE policy. Further information on these datasets is available via the HMS LINCS datasbase (see datasets 20324, 20325, and 20349) and Synapse (syn13856689). Data analysis software are available on Github.
The following data are available for download; imaging data are available on request.
| Manuscript Figure/Supplemental Figure | Associated Dataset | Synapse ID | HMS LINCS Dataset |
|---|---|---|---|
| 1 | Cardiotoxic phenotypes induced by four TKIs (Supplementary Table 1) | syn18458239 | |
| Pharmacological properties of four TKIs (Supplementary Table 2) | syn18458240 | ||
| 2, 3, S2, S3, S4, S6, S7 | RNA-seq gene expression data | syn13908807 | 20324 |
| 2, S2, S3 | qRT-PCR validation data | syn13905466 | 20325 |
| Genes for qRT-PCR validation (Supplementary Table 3) | syn18458241 | ||
| 3, S4 | Genes in RNA-seq clusters with different enriched GO terms (Supplementary Table 4) | syn18458242 | |
| 4, 5, S5, S6, S7 | Proteomics data | syn13905629 | 20349 |
| 4, S5 | Protein clusters with different enriched GO terms (Supplementary Table 5) | syn18458243 | |
| 5, S6, S7 | Kinases expressed by Cor.4U hiPSC-CMs (Supplementary Table 6) | syn18458244 | |
| Detection of TKI target mRNA or protein in Cor.4U hiPSC-CM cells (Supplementary Table 7) | syn18458245 | ||
| 6 | Metabolic profile of Cor.4U hiPSC-CMs following treatment with Sorafenib (Supplementary Table 8) | syn18458247 |
Funding Sources
The research was supported by FDA contract HHSF223201400052C and NIH grants U54-CA225088 and U54-HL127365 to PKS, and AHA fellowship 15POST25230014 and NIH F32 HL14223 to HW.