Publication Summary
Encoding growth factor identity in the temporal dynamics of a transcription factor under combinatorial regulation
Somponnat Sampattavanich1,2, Bernhard Steiert1,3,4, Bernhard A. Kramer1,5, Benjamin M. Gyori1, John G. Albeck6, and Peter K. Sorger1
1HMS LINCS Center and Laboratory of Systems Pharmacology, Department of Systems Biology, Harvard Medical School, 200 Longwood Ave, Boston, USA; 2Siriraj Laboratory for Systems Pharmacology, Department of Pharmacology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand; 3Institute of Physics, University of Freiburg, Freiburg, Germany; 4Freiburg Center for Systems Biology, University of Freiburg, Freiburg, Germany; 5Division of Systems Biology of Signal Transduction, German Cancer Research Center, Heidelberg, Germany; 6Department of Molecular and Cellular Biology, University of California, Davis, USA
doi:10.1016/j.cels.2018.05.004
Synopsis
The Forkhead box O3 transcription factor (FoxO3) functions as an integrative node for diverse upstream signaling networks, and has been implicated in a number of biological processes including cycle arrest, apoptosis, oxidative stress, cell migration and cell metabolism. Regulators of FoxO3, including phosphorylation, energy stress, genotoxic stress, and cytokines, control its abundance, subcellular localization and DNA-binding capacity. In this paper we study how the identities and concentrations of growth factors are encoded in the dynamics of FoxO3 activation. Transcription factors like FoxO3 often switch between on and off states repeatedly over the course of a 12-24 hours period. Immunoblotting and similar methods can obscure this switching behavior due to asynchronicity between cells. In contrast, our use of time-lapse microscopy of fluorescent reporter proteins reveals that FoxO3 exhibits complex low wavelength and pulsatile translocation dynamics responsive to combinatorial control by ERK and Akt with the potential to encode the identities and concentrations of multiple extracellular growth factors.
PREVIOUS: The human FoxO3 transcription factor exhibits complex low wavelength and pulsatile translocation dynamics responsive to combinatorial control by ERK and Akt with the potential to encode the identities and concentrations of multiple extracellular growth factors.
Key Findings
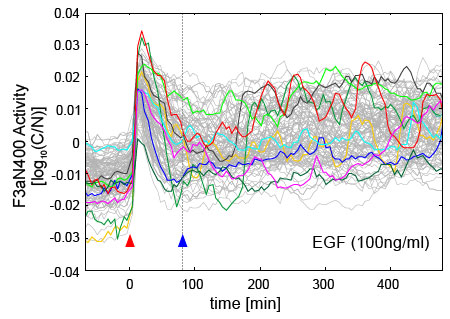
Figure 1D: Single-cell translocation trajectories of the F3aN400-Venus reporter, a proxy for FoxO3, in 184A1 cells following exposure to 100 ng/ml EGF. Cells were imaged every 5 minutes with >100 trajectories shown; 10 are randomly highlighted in color. Red arrowhead marks the time of ligand addition, denoted as t=0. Translocation trajectories are synchronous up to t=80 minutes (blue arrowhead) after which asynchronous pulsing predominates.
- FoxO3 nuclear-cytosolic shuttling is pulsatile but not oscillatory.
- FoxO3 dynamics can encode growth factor identities.
- ERK and Akt control FoxO3 dynamics in a combinatorial manner.
- Diversity of FoxO3 states is lost in cancer.
Abstract
Extracellular ligands signal to transcription factors via a limited number of cytoplasmic kinase cascades. It remains unclear how such cascades encode ligand identity and concentration. In this paper we use live-cell imaging and statistical modeling to study FoxO3, a transcription factor under combinatorial control that regulates diverse aspects of cellular physiology. We show that FoxO3 nuclear-to-cytosolic translocation has two temporally and qualitatively distinct phases that vary in magnitude with growth factor identity and cell type. These phases are differentially regulated by Akt and ERK and comprise synchronous translocation soon after ligand addition followed by an extended period of back and forth shuttling (pulsing). FoxO3 pulsing does not have a characteristic frequency, unlike a true oscillator, but early and late dynamics have low mutual information and can encode different information. The diversity of FoxO3 states is lost in cancer cells, however, in which ERK and Akt are dysregulated by oncogenic mutation.
Explore the data
| IGF treatment (weak FoxO3 reporter pulsing) | |
| BTC treatment (strong pulsing) | |
| BTC plus Akt inhibitor (pulsing reduced) |
We encourage readers to explore the findings and the data underlying this study. Through the links below, readers can download all single-cell and population data, as well as fitted parameter data, for key figures from the manuscript.
Available data and resources
| Data | Figure 1D/2A | Single-cell reporter measurements | Details | Download (.zip) |
| Single-cell reporter pulsing metrics | Details | Download (.zip) | ||
| Mean reporter pulsing metrics | Details | Download (.zip) | ||
| Original raw images | Details | Download | ||
| Data | Figure 5A | Single-cell reporter measurements | Details | Download (.zip) |
| Single-cell reporter pulsing metrics | Details | Download (.zip) | ||
| Mean reporter pulsing metrics | Details | Download (.zip) | ||
| Original raw images | Details | Download | ||
| Data | Figure 5B | Single-cell reporter measurements | Details | Download (.zip) |
| Single-cell reporter pulsing metrics | Details | Download (.zip) | ||
| Mean reporter pulsing metrics | Details | Download (.zip) | ||
| Original raw images | Details | Download | ||
| Data | Figure 6B | Single-cell reporter measurements | Details | Download |
| Single-cell reporter pulsing metrics | Details | Download | ||
| Mean reporter pulsing metrics | Details | Download | ||
| Original raw images | Details | Download | ||
| Data | Figure 7B | Single-cell reporter measurements | Details | Download |
| Single-cell reporter pulsing metrics | Details | Download | ||
| Mean reporter pulsing metrics | Details | Download | ||
| Original raw images | Details | Download | ||
| Software | All code to reproduce the figures from the data is available at GitHub | Details | Download (.zip) | |
| Data | Intermediate data for use with the figure generation code | Details | ||
Grayed-out entries are currently in preparation.
Funding sources
NIH grants P50 GM107618 and U54 HL127365, a “Chalermphrakiat” Grant from the Faculty of Medicine at Siriraj Hospital, Mahidol University and the Thai Research Fund (TRG5880094), and the German Ministry of Education and Research (SBEpo 0316182A and 0316042G)