Publication Summary
Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE)
Evan O. Paull1, Daniel E. Carlin1, Mario Niepel2, Peter K. Sorger2, David Haussler1,3, and Joshua M. Stuart1
1 Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, CA; 2 HMS LINCS Center, Harvard Medical School, Boston, MA; 3 Howard Hughes Medical Institute, Santa Cruz, CA
Bioinformatics (2013) 29 (21), 2757-2764.
doi:10.1093/bioinformatics/btt471 / PMID:23986566 / PMCID:PMC3799471
Synopsis
Identifying the cellular wiring that connects genomic perturbations to transcriptional changes in cancer is essential to gain a mechanistic understanding of disease initiation and progression and ultimately to predict drug response. We have developed a method called Tied Diffusion Through Interacting Events (TieDIE) that uses a network diffusion approach to connect genomic perturbations to gene expression changes characteristic of cancer subtypes.
Key findings
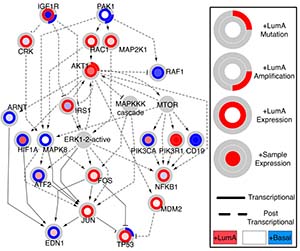
- TieDIE computes a subnetwork of protein–protein interactions, predicted transcription factor-to-target connections, and curated interactions from literature that connects genomic and transcriptomic perturbations.
- When TieDIE was applied to The Cancer Genome Atlas and a breast cancer cell line dataset, it identified key signaling pathways, with examples impinging on MYC activity.
- TieDIE finds interlinking genes that are predicted to correspond to essential components of cancer signaling and may provide a mechanistic explanation of tumor character and suggest subtype-specific drug targets.
- Using TieDIE we also are able to infer networks that integrate multi-modal data from a single tumor sample with prior pathway knowledge. These networks may suggest mechanisms of drug resistance or sensitivity for a given patient.
Available data and software
| Data | Genes in TieDIE network inferred with TCGA data | Download (.xlsx) | |
| Data | Genes in the TieDIE networks inferred with Cell Line Data and TCGA data | Download (.xlsx) | |
| Data | Genes in the TieDIE network inferred for a single Luminal A sample | Download (.xlsx) | |
| Software | TieDIE source code, a tutorial, and further information about the software available through the TieDIE website hosted by the UCSC Systems Biology Department | Details | TieDIE at GitHub |
Funding sources
NIH LINCS grant U54 HG006097 to PKS; National Cancer Institute TCGA grant 5U24CA143858 to JMS and DH; National Science Foundation CAREER Award DBI 0845783 to JMS.