Publication Summary
Analysis of growth factor signaling in genetically diverse breast cancer lines
Mario Niepel1*, Marc Hafner1*, Emily A. Pace2*, Mirra Chung1, Diana H. Chai2, Lili Zhou1, Jeremy L. Muhlich1, Birgit Schoeberl2, and Peter K. Sorger1
1 HMS LINCS Center, Harvard Medical School, Boston, MA; 2 Merrimack Pharmaceuticals, Cambridge, MA
BMC Biol (2014) 12:20.
doi:10.1186/1741-7007-12-20 / PMID:24655548 / PMCID:PMC4234128
Synopsis
Growth factor signaling is a key determinant of cellular behavior in normal and diseased tissue. Here, we determined the variance of signaling responses across a panel of breast cancer lines and identified potential molecular underpinnings of these responses. We correlated basal receptor levels with ligand responses to derive a simple heuristic that approximates the responsiveness of cells to individual growth factors.
Key findings
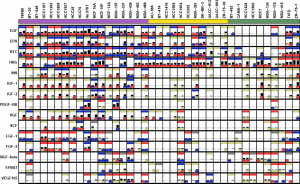
- By analyzing the response at 3 time points of 39 breast cancer cell lines to 15 ligands at 2 doses at the level of pERK and pAKT levels and relating these responses to clinical cancer subtypes and the expression and phosphorylation levels of RTKs, we found that the response to ErbB ligands is ubiquitous across virtually all cell lines and many respond to HGF, FGFs, and IGF/INS.
- TNBCs are highly responsive to a variety of ligands, whereas HER2amp cell lines only show muted responses.
- The responsiveness of individual cell lines can be determined by a simple heuristic.
Abstract
Background Soluble growth factors present in the microenvironment play a major role in tumor development, invasion, metastasis, and responsiveness to targeted therapies. While the biochemistry of growth factor-dependent signal transduction has been studied extensively in individual cell types, relatively little systematic data are available across genetically diverse cell lines.
Results We describe a quantitative and comparative dataset focused on immediate-early signaling that regulates the AKT (AKT1/2/3) and ERK (MAPK1/3) pathways in a canonical panel of well-characterized breast cancer lines. We also provide interactive web-based tools to facilitate follow-on analysis of the data. Our findings show that breast cancers are diverse with respect to ligand sensitivity and signaling biochemistry. Surprisingly, triple negative breast cancers (TNBCs; which express low levels of ErbB2, progesterone and estrogen receptors) are the most broadly responsive to growth factors and HER2amp cancers (which overexpress ErbB2) the least. The ratio of ERK to AKT activation varies with ligand and subtype, with a systematic bias in favor of ERK in hormone receptor positive (HR+) cells. The factors that correlate with growth factor responsiveness depend on whether fold-change or absolute activity is considered the key biological variable, and they differ between ERK and AKT pathways.
Conclusions Responses to growth factors are highly diverse across breast cancer cell lines, even within the same subtype. A simple four-part heuristic suggests that diversity arises from variation in receptor abundance, an ERK/AKT bias that depends on ligand identity, a set of factors common to all receptors that varies in abundance or activity with cell line, and an “indirect negative regulation” by ErbB2. This analysis sets the stage for the development of a mechanistic and predictive model of growth factor signaling in diverse cancer lines.
Explore the data
We encourage readers to explore the findings and the data underlying this study through the associated project exploration website, which presents the data through several customized visualization tools and allows readers to browse the data by cell line and by ligand. Through the links below, readers also can access the original datasets and the results of some of the presented analyses, as well as the software for the signaling data matrix.
Available data and software
| Data | All basal levels (HMS Dataset #20137 from Niepel et al (2013). Sci Signal. 6, ra84). | Details | Download (.xlsx) |
| Data | All ligand responses (HMS Dataset #20140 from Niepel et al (2013). Sci Signal. 6, ra84). | Details | Download (.xlsx) |
| Signatures | Results of kinetic clustering and dose-dependence classification. | Details | Download (.xlsx) |
| Software | A link to our HMS LINCS GitHub for access to the code for the signaling data matrix presented on the associated project exploration website. | hmslincs at GitHub |
Funding sources
NIH LINCS grant U54 HG006097; NIH grant U54 CA112967 (for access to ICBP43 cells); and fellowships from the Swiss National Science Foundation (PBELP3_140652 and P300P3_147876) to MH.