Project Summary
Single-cell analysis of subcellular morphology in response to perturbagen treatment of breast cancer cells
Caitlin E. Mills1,2, David W. Andrews1, and Peter K. Sorger2
1 HMS LINCS Center, Harvard Medical School, Boston, MA; 2 Department of Biochemistry, University of Toronto, Toronto, Canada
(This dataset is unpublished. Please see our terms of use.)
Synopsis
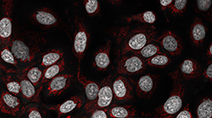 Kinase inhibitors are important candidates for targeted therapy for breast cancer. In this unpublished dataset, five breast cancer cell lines and one non-tumorigenic breast cell line were treated with a panel of 105 kinase inhibitors. Cells were stained with nuclear and mitochondrial dyes and imaged live following 24 hours of drug exposure to detect early signs of cell stress.
Kinase inhibitors are important candidates for targeted therapy for breast cancer. In this unpublished dataset, five breast cancer cell lines and one non-tumorigenic breast cell line were treated with a panel of 105 kinase inhibitors. Cells were stained with nuclear and mitochondrial dyes and imaged live following 24 hours of drug exposure to detect early signs of cell stress.
Explore the data
We encourage readers to explore and make use of the data underlying this study through the links in the table below. We provide access to the complete set of feature recordings from 1.6 million individual cells across 6 cell lines and 3 doses of 105 kinase inhibitors as well as access to the full set of raw image data through our OMERO server. The feature extraction script and parameter files used in this study are provided below and described in greater detail on the Andrews lab website. We also provide a downloadable file that describes each of the morphological features that was assayed. Please see Haralick et al (1973)1 and Hamilton et al (2007)2 for additional details about the Haralick and TAS texture features specifically.
Available data and software
| Data | Sample dataset containing selected feature measurements for all cell lines treated with 2 perturbagens (HMS Dataset #20244). | Download (.xls) |
| Data | Complete dataset containing all feature measurements across all cell lines and perturbagens. | Download replicate #1 (.zip) |
| Download replicate #2 (.zip) | ||
| Download replicate #3 (.zip) | ||
| Data | Complete set of raw image data across all cell lines and perturbagens (via our OMERO server). | View data |
| Software | Complete set of feature extraction script and parameter files used in this study. | Download (.zip) |
| Related Information | Detailed descriptions of each measured feature. | Download (.xlsx) |
Funding sources
NIH LINCS grant U54 HG006097 and U54 HL127365 to PKS and CIHR grant FRN 10490 to DWA.
References
1. Haralick, R.M., Shanmugam, K., and Dinstein, I. (1973) Textural features for image classification. IEEE Trans Syst Man Cybern. SMC-3, 610-621.
2. Hamilton, N.A. Pantelic, R.S., Hanson, K., and Teasdale, R.D. (2007) Fast automated cell phenotype image classification. BMC Bioinformatics. 8, 110. doi:10.1186/1471-2105-8-110 PMID:17394669 PMCID:PMC1847687