Publication Summary
Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state
Mohammad Fallahi-Sichani1, Verena Becker1, Benjamin Izar2,3, Gregory J. Baker1, Jia-Ren Lin4, Sarah A. Boswell1, Parin Shah2, Asaf Rotem2, Levi A. Garraway2,3,5, and Peter K. Sorger1,4,5
1Program in Therapeutic Sciences, Department of Systems Biology, Harvard Medical School, Boston, MA; 2 Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA; 3 Broad Institute of Harvard and MIT, Cambridge, MA; 4 HMS LINCS Center and Laboratory of Systems Pharmacology, Harvard Medical School, Boston, MA; 5 Ludwig Center at Harvard, Harvard Medical School, Boston, MA
Mol Syst Biol (2017) 13(1): 905
doi:10.15252/msb.20166796 / PMID:28069687 / PMCID:PMC5248573
Synopsis
Single-cell imaging, transcriptional profiling, and biochemical profiling of the responses of BRAFV600E melanoma cells to Vemurafenib uncovered a slowly dividing, de-differentiated cell state that is associated with drug resistance but that is inhibitable by novel drug combinations.
Key Findings
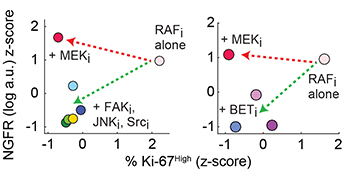
- Cell-to-cell variability in BRAFV600E melanomas generates drug-tolerant subpopulations.
- The drug-tolerant, slowly-dividing NFGRHigh state is transiently heritable.
- Drugs against a proposed c-Jun/ECM/FAK/Src cascade block acquisition of this phenotype.
- The NGFRHigh drug-tolerant state is also blocked by BET inhibitors in vitro and in vivo.
- Drugs that block adaptation by cell subpopulations increase cell killing by RAF/MEK inhibitors.
Abstract
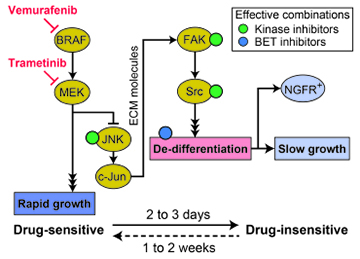 Treatment of BRAF-mutant melanomas with MAP-kinase pathway inhibitors is paradigmatic of the promise of precision cancer therapy but also highlights problems with drug resistance that limit patient benefit. We use live-cell imaging, single-cell analysis and molecular profiling to show that exposure of tumor cells to RAF/MEK inhibitors elicits a heterogeneous response in which some cells die, some arrest and the remainder adapt to drug. Drug-adapted cells up-regulate markers of the neural crest (e.g. NGFR), a melanocyte precursor, and grow slowly. This phenotype is transiently stable, reverting to the drug-naïve state within 9 days of drug withdrawal. Transcriptional profiling of cell lines and human tumors implicates a c-Jun/ECM/FAK/Src cascade in de-differentiation in about one-third of cell lines studied; drug-induced changes in c-Jun and NGFR levels are also observed in xenograft and human tumors. Drugs targeting the c-Jun/ECM/FAK/Src cascade as well as BET bromodomain inhibitors increase the maximum effect (Emax) of RAF/MEK kinase inhibitors by promoting cell killing. Thus, analysis of reversible drug resistance at a single-cell level identifies signaling pathways and inhibitory drugs missed by assays that focus on cell populations.
Treatment of BRAF-mutant melanomas with MAP-kinase pathway inhibitors is paradigmatic of the promise of precision cancer therapy but also highlights problems with drug resistance that limit patient benefit. We use live-cell imaging, single-cell analysis and molecular profiling to show that exposure of tumor cells to RAF/MEK inhibitors elicits a heterogeneous response in which some cells die, some arrest and the remainder adapt to drug. Drug-adapted cells up-regulate markers of the neural crest (e.g. NGFR), a melanocyte precursor, and grow slowly. This phenotype is transiently stable, reverting to the drug-naïve state within 9 days of drug withdrawal. Transcriptional profiling of cell lines and human tumors implicates a c-Jun/ECM/FAK/Src cascade in de-differentiation in about one-third of cell lines studied; drug-induced changes in c-Jun and NGFR levels are also observed in xenograft and human tumors. Drugs targeting the c-Jun/ECM/FAK/Src cascade as well as BET bromodomain inhibitors increase the maximum effect (Emax) of RAF/MEK kinase inhibitors by promoting cell killing. Thus, analysis of reversible drug resistance at a single-cell level identifies signaling pathways and inhibitory drugs missed by assays that focus on cell populations.
Explore the data
We encourage readers to explore the datasets associated with this paper, including mining them for additional insights. All total, six datasets are publicly available through the HMS LINCS Database via the links below. Also available for download is a table summarizing the protein target, fluorophore, and dilution details for each of the antibody reagents used in HMS Datasets #20274, 20275, 20276, and 20277.
Available Data and Resources
| Data | Viability and apoptosis measured by imaging in BRAF(V600E/D) melanoma cell lines following treatment with combinations of two compounds (HMS Dataset #20272) | Details | Download (.xls) |
| Data | Viability and apoptosis measured by imaging in BRAF(V600E/D) melanoma cell lines following treatment with combinations of three compounds (HMS Dataset #20273) | Details | Download (.xls) |
| Data | Phosphorylation state and protein levels measured by imaging in BRAF(V600E/D) melanoma cell lines following treatment with combinations of two compounds (HMS Dataset #20274) | Details | Download (.xls) |
| Data | Phosphorylation state and protein levels measured by imaging in BRAF(V600E/D) melanoma cell lines following treatment with combinations of two compounds (HMS Dataset #20275) | Details | Download (.xls) |
| Data | Protein levels measured by imaging in BRAF(V600E/D) melanoma cell lines following treatment with Vemurafenib in combination with chromatin-targeting compounds (HMS Dataset #20276) | Details | Download (.xls) |
| Data | Phosphorylation state and protein levels measured by imaging in BRAF(V600E/D) melanoma cell lines following treatment with combinations of two compounds (HMS Dataset #20277) | Details | Download (.xls) |
| Reagent Metadata | Antibody summary table (for HMS Datasets #20274, 20275, 20276, and 20277) | Download (.xlsx) |
Funding Sources
NIH grants U54 HL127365 (LINCS), CA139980 and GM107618 to PKS, NIH grant K99CA194163 to MF-S, a Merck Fellowship of the Life Sciences Research Foundation to MF-S, grants from the Adelson Medical Research Foundation and the Melanoma Research Alliance to LAG, a grant from the Ludwig Center at Harvard to PKS and LAG, and a grant from the DFCI Wong Family Award to BI.