About the HMS LINCS Center
The Harvard Medical School (HMS) LINCS Center is funded by NIH grant U54 HL127365 and is part of the NIH Library of Integrated Network-based Cellular Signatures (LINCS) Program. The overall goals of this program are to collect and disseminate data and analytical tools needed to understand how human cells respond to perturbation by drugs, the environment, and mutation. Further information about LINCS and other participating Centers is available at the program website.
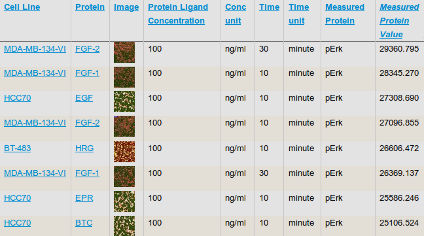
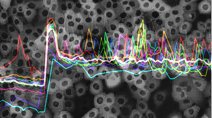
The HMS LINCS Center aims to discover fundamental principals of cellular response to perturbation including the relationship between dose and response, the origin and significance of cell-to-cell variation, and the molecular basis of drug sensitivity and resistance. Data generated at HMS include multiplex biochemical, proteomic, and imaging assays for which dissemination standards are poorly developed; improving these and liberating all data, algorithms, and conclusions from our notebooks and papers is a key goal of the Center. The Center also develops new algorithms for large-scale analysis of complex perturbagen-response datasets, which are available through customized project summary pages. We typically publish at least one biological analysis of each dataset, but these papers describe only one of many uses for the data. We encourage users to explore the primary data based on their own ideas and algorithms.
Assays
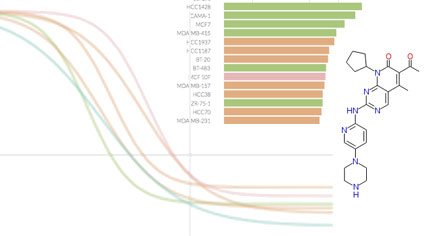
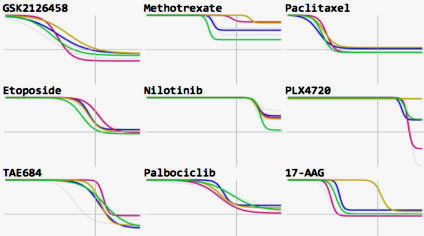
We focus on biochemical and cell-based assays on (i) small molecule kinase inhibitors, a leading class of therapeutic agents for the treatment of cancer, autoimmune diseases, and other diseases; (ii) epigenome modifiers such as bromodomain and HDAC inhibitors; and (iii) naturally occurring ligands such as growth factors and inflammatory cytokines. Overall we are profiling the responses of ~100 cell lines and primary cell types to ~400 perturbagens across dose and time.
Access to data, algorithms, and tools
HMS LINCS provides timely public access to all relevant data and software in accordance with the data release policy of the NIH LINCS program. Simple download and programmatic access are both supported.
Data exploration and use cases: Summary and exploration pages for HMS LINCS publications provide descriptions of key findings, links to relevant datasets in the HMS LINCS Database, and custom data visualization tools. These and other tools are available via our software page.
Data access: All publicly-released HMS LINCS datasets and reagent metadata are accessible through the HMS LINCS Database, where search functions make it possible to query for specific datasets and reagents (e.g. by cell type or perturbagen). These search and download functions are actively being improved, and we encourage users to send feedback to lincs-feedback [at] hms.harvard.edu.