Publication Summary
Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes
Jia-Ren Lin * 1,2, Benjamin Izar * 1,2,3,4, Shu Wang 1, Clarence Yapp 1, Shaolin Mei 1,3, Parin Shah 3, Sandro Santagata 1,2,5,6 and Peter K Sorger 1,2
* These authors contributed equally to this work.
1 Laboratory of Systems Pharmacology, Harvard Medical School, Boston, MA
2 Ludwig Center for Cancer Research at Harvard, Harvard Medical School, Boston, MA
3 Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA
4 Broad Institute of MIT and Harvard, Cambridge, MA
5 Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA
6 Department of Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA
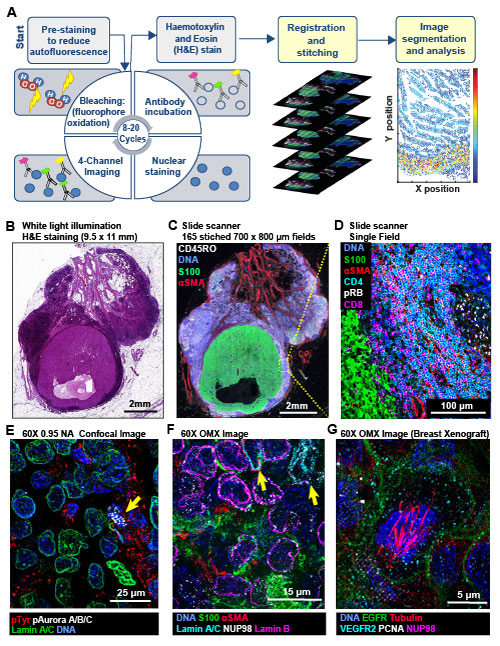
Multi-scale imaging of t-CyCIF specimens. (A) Schematic of the cyclic process whereby t-CyCIF images are assembled via multiple rounds of four-color imaging. (B) Bright-field H&E image of a metastasectomy specimen that includes a large metastatic melanoma lesion and adjacent benign tissue. The H&E staining was performed after the same specimen had undergone t-CyCIF. (C) Representative t-CyCIF staining of the specimen shown in (A) stitched together using the Ashlar software from 165 successive CyteFinder fields using a 20X/0.8NA objective (D) One field from (B) at the tumor-normal junction demonstrating staining for S100-postive malignant cells, α-SMA positive stroma, T lymphocytes (positive for CD3, CD4 and CD8), and the proliferation marker phospho-RB (pRB). (E) A melanoma imaged on a GE INCell Analyzer 6000 confocal microscope to demonstrate sub-cellular and sub-organelle structures. (F) Staining of a melanoma sample using the OMX Blaze structured illumination microscope with a 60x/1.42NA objective. (G) Staining of a patient-derived mouse xenograft breast tumor using the OMX Blaze showing a spindle in a mitotic cell.
Synopsis
Tissue-based cyclic immunofluorescent microscopy (t-CyCIF) is a simple method for generating highly multiplexed optical images from formalin-fixed paraffin-embedded (FFPE) tissue samples routinely used for histopathological diagnosis of human disease. The method is based on previously described single-cell imaging approaches and readily implemented on existing instruments (Gerdes et al. 2013, Lin et al. 2015, 2016).
Key Findings
- t-CyCIF produces highly multiplexed images (of up to 60 antigens) of normal and diseased tissue using an iterative process in which low-plex fluorescence images are repeatedly collected from the same tissue sample and then assembled into a high dimensional representation.
- t-CyCIF uses widely available reagents and conventional microscopes and can be implemented inexpensively in most research and clinical laboratories. Super-resolution imaging has been demonstrated for multiple samples.
- To date over 200 commercial antibodies have been tested for their compatibility with t-CyCIF. We are working to develop a generally useful antibody validation resource. More information is available at www.cycif.org, a community resource with regularly updated antibody lists, protocols, and data.
Abstract
The architecture of normal and diseased tissues strongly influences the development and progression of disease as well as responsiveness and resistance to therapy. We describe a tissue-based cyclic immunofluorescence (t-CyCIF) method for highly multiplexed immuno-fluorescence imaging of formalin-fixed, paraffin-embedded (FFPE) specimens mounted on glass slides, the most widely used specimens for histopathological diagnosis of cancer and other diseases. t-CyCIF generates up to 60-plex images using an iterative process (a cycle) in which conventional low-plex fluorescence images are repeatedly collected from the same sample and then assembled into a high dimensional representation. t-CyCIF requires no specialized instruments or reagents and is compatible with super-resolution imaging; we demonstrate its application to quantifying signal transduction cascades, tumor antigens and immune markers in diverse tissues and tumors. The simplicity and adaptability of t-CyCIF makes it an effective method for pre-clinical and clinical research and a natural complement to single-cell genomics.
Available data and software
Links to primary data are listed below by figure and panel number. The data obtained from experiments that test antibody order of addition (show in Figures 5 & 6) are available as a Jupyter notebook. A list of validated antibodies is also available for download
t-CyCIF specimens can be up to several square centimeters in area. Imaging these specimens at subcellular resolution is accomplished by stitching many succesive fields together. The final reconstructions often comprise 100-200 individual fields each with 20-60 channels. Methods for sharing such data are not fully developed. All individual image panels can be obtained from the HMS OMERO image database via links listed below. Large-scale stitched and registered image panels can be obtained at www.cycif.org (RRID:SCR_016267). We are also working with the Image Data Resource (IDR) to make t-CyCIF images available via download. This web page will be updated with these links as they become available.
| Figure | View original image tiles | View stitched mosaic images | Download figures or source data |
|---|---|---|---|
| 1A | Figure 1A pdf | ||
| 2E | Melanoma sample | ||
| 2F | Patient-derived mouse xenograft breast tumor | ||
| 3 | Tonsil composites | ||
| 4B | Histogram single-cell source data | ||
| 4C | Bleaching timecourse | ||
| 4E | Cell loss through cycles | ||
| 5 & 6 | 16-cycle stitched images (in preparation) | Tonsil A mosaic Tonsil B mosaic |
Single-cell data |
| 6 | Jupyter notebook (non-interactive preview) | ||
| 7 & 8 | PDAC composites | PDAC mosaic | Single-cell data |
| 7E | Single-cell intensity heatmap | ||
| 8 A, C & D | Single-cell data FCS file | ||
| 9 | RCC composites | Single-cell data | |
| 9C | Immune cell counts | ||
| 10 | TMA panel composites | TMA mosaic | Single-cell data |
| 11 & 12 | GBM composites | GBM mosaic | Single-cell data |
| 11C | Normalized Entropy for EGFR, pERK & pRB | ||
| 12D | Ratios of EMGM clusters in different regions |
Associated code:
Third-party software:
Funding Sources
NIH grants U54-HL127365 (LINCS), P50-GM107618, and R41-CA224503 to PKS and a DF/HCC GI SPORE Developmental Research Project Award (BI), DFCI Claudia Adams Barr Program for Innovative Cancer Research Award grant K08CA222663 to BI.
Related References
1. Gerdes, M. J. et al. (2013) Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. PNAS. 110(29):11982-7. doi:10.1073/pnas.1300136110 PMID:23818604 PMCID:PMC3718135
2. Lin, J.-R., Fallahi-Sichani, M., and Sorger, P.K. (2015) Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 6:8390. doi:10.1038/ncomms9390 PMID:26399630 PMCID:PMC4587398
3. Lin, J.R., Fallahi-Sichani, M., Chen, J.Y., and Sorger, P.K. (2016) Cyclic Immunofluorescence (CycIF), a highly multiplexed method for single-cell imaging. Curr Protoc Chem Biol. 8(4):251-264. doi:10.1002/cpch.14 PMID:27925668 PMCID:PMC5233430