Project Summary
Dynamics of perturbagen responses in living single cells
Somponnat Sampattavanich1,2 and Peter K. Sorger1
1 HMS LINCS Center, Harvard Medical School, Boston, MA; 2 Current address: Department of Pharmacology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
(This dataset is unpublished. Please see our terms of use.)
Synopsis
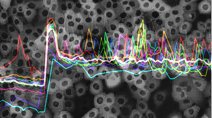 Many signal transduction networks exhibit pulsatile behavior whose amplitude and dynamics regulate downstream processes (see Levine and Elowitz (2013)). In this unpublished dataset, human mammary epithelial TCT cells expressing a FRET-based reporter of ERK activity were treated with EGF and various clinical grade inhibitors of ErbB signal transduction that have fundamentally different chemistry. Gefitinib binds reversibly to the active kinase conformation, Lapatinib binds ‘irreversibly’ to the inactive kinase conformation (that is, it dissociates very slowly), and Canertinib is a covalent modifier.
Many signal transduction networks exhibit pulsatile behavior whose amplitude and dynamics regulate downstream processes (see Levine and Elowitz (2013)). In this unpublished dataset, human mammary epithelial TCT cells expressing a FRET-based reporter of ERK activity were treated with EGF and various clinical grade inhibitors of ErbB signal transduction that have fundamentally different chemistry. Gefitinib binds reversibly to the active kinase conformation, Lapatinib binds ‘irreversibly’ to the inactive kinase conformation (that is, it dissociates very slowly), and Canertinib is a covalent modifier.
Explore the data
We encourage readers to explore the findings and the data underlying this study through the links in the table below and through the associated project exploration website, which presents an interactive lookup table through which users can access computed ERK trajectories, movies of cells showing ERK activity across each condition, and recordings of the position and ERK signal across time for each cell analyzed. Image segmentation information and access to the full set of raw image data via our OMERO server also is provided.
Available data and software
| Data | Interactive lookup table across all conditions that provides access to computed trajectory plots, movies of cells showing ERK activity, and a .csv file of the position and ERK signal across time for each cell analyzed. | View table |
| Data | Computed ERK trajectories with image segmentation information. | Download (2.5 GB zip) |
| Data | Raw image data via our OMERO server. | View data |
| Software | A link to our HMS LINCS GitHub for access to the code for the interactive lookup table presented on the associated project exploration website. | hmslincs at GitHub |
Funding sources
NIH LINCS grant U54 HG006097 and U54 HL127365 to PKS.