Publication Summary
Fractional killing arises from cell-to-cell variability in overcoming a caspase activity threshold
Jérémie Roux1,3, Marc Hafner1,3, Samuel Bandara1, Joshua J. Sims1, Hannah Hudson2, Diana Chai2, and Peter K. Sorger1
1 Department of Systems Biology, Harvard Medical School, Boston, MA; 2 Merrimack Pharmaceuticals, Cambridge, MA; 3 JR and MH contributed equally to this work.
Mol Syst Biol (2015) 11: 803.
doi:10.15252/msb.20145584 / PMID:25953765 / PMCID:PMC4461398
Synopsis
Non-genetic cell-to-cell variability results in fractional killing by TRAIL and therapeutic antibody agonists, limiting their effectiveness as anti-cancer drugs. A simple model of initiator caspase dynamics reveals a threshold in caspase activity that separates dying and surviving cells.
Key findings
- A model of initiator caspase activity is predictive of fractional killing by TRAIL.
- The model identifies a caspase activity threshold that is constant across dose and type of agonist.
- Therapeutic antibodies targeting TRAIL receptors kill poorly because this threshold is not crossed.
- The caspase activity threshold and classic MOMP threshold interact to determine cellular sensitivity to apoptosis inducers.
Abstract
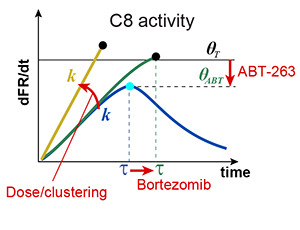
When cells are exposed to death ligands such as TRAIL, a fraction undergoes apoptosis and a fraction survives; if surviving cells are re-exposed to TRAIL, fractional killing is once again observed. Therapeutic antibodies directed against TRAIL receptors also cause fractional killing, even at saturating concentrations, limiting their effectiveness. Fractional killing arises from cell-to-cell fluctuations in protein levels (extrinsic noise), but how this results in a clean bifurcation between life and death remains unclear. In this paper we identify a threshold in the rate and timing of initiator caspase activation that distinguishes cells that live from those that die; by mapping this threshold we can predict fractional killing of cells exposed to natural and synthetic receptor agonists alone or in combination with sensitizing drugs such as bortezomib. A phenomenological model of the threshold also quantifies the contributions of two resistance genes (c-FLIP and Bcl-2), providing new insight into the control of cell fate by opposing pro-death and pro-survival proteins and suggesting new criteria for evaluating the efficacy of therapeutic TRAIL receptor agonists.
Explore the data
We encourage readers to explore the findings and the data underlying this study through the associated project exploration website, which presents plots of the smoothed single-cell trajectories and fitted parameters for each replicate of each individual treatment condition. Through the links below and on the interactive website, readers also can download all single-cell trajectory data and fitted parameter data for independent analysis.
Available data, signatures and software
| Data and signatures | All single-cell raw data, smoothed trajectories, and fitted parameters for each replicate for each treatment condition. | Download (.zip) | |
| Signatures | Well-by-well summary of the fitted parameters for each replicate for each treatment condition (HMS Dataset #20232). | Details | Download (.tsv) |
| Data and signatures | Population-based viability and C8 activity results for Apomab, Apomab + anti-Fc, and TRAIL treatments in multiple cell lines. | Download (.zip) | |
| Software | The MATLAB code for the image processing routines developed for this project. | Download (.zip) |
Funding sources
NIH LINCS grant U54 HL127365 and grant CA139980 to PKS; a Swiss National Science Foundation fellowship (PBELP3_140652 and P300P3_147876) to MH; and a Marie Curie International Incoming Fellowship within the 7th European Community Framework Programme (proposal SysBioDRez, N° 626190, call reference: FP7-PEOPLE-2013-IIF) to JR.